Colloidal Gold Nanoparticles Induce Apoptosis in MCF-7 Human Breast Adenocarcinoma Cells
Adewumi H, Carter GÂ and Bhuiyan S*
Department of Arts and Sciences, Jarvis Christian College, U.S. Highway 80 East PR 7631, Hawkins, Texas, USA
- Corresponding Author:
- Bhuiyan S
Department of Arts and Sciences
Jarvis Christian College, U.S. Highway 80 East PR 7631
Hawkins, Texas, USA
Tel: +19033728052
E-mail: sbhuiyan@jarvis.edu
Received: November 04, 2019; Accepted: November 28, 2019; Published: December 06, 2019
Citation:Adewumi H, Carter G, Bhuiyan S (2019) Colloidal Gold Nanoparticles Induce Apoptosis in MCF-7 Human Breast Adenocarcinoma Cells. Nano Res Appl Vol.5 No.2:5
Abstract
Nanoparticles have been widely used as remedies for disorders for a long time. They are 10-9 m specks of substances that can be found both naturally and synthesized in the laboratory with metal and nonmetal materials. In this study, gold nanoparticles (AuNPs) were synthesized using the citrate reduction method, and the 35 nm size of the nanoparticles was determined using a UV-Vis Spectrophotometer at 525 nm wavelength. The synthesized nanoparticles were further studied on MCF-7 breast cancer cells to understand how various genes are expressed in the induction of apoptosis in signal transduction pathways. The results obtained from the anticancer activity of the gold nanoparticles showed approximately 90% inhibition of cell growth after 72 hours of treatment. Western blot analysis demonstrated the downregulation of p44/42 MAPK (ERK1/2) protein due to gold nanoparticle treatment. Moreover, reverse transcription-polymerase chain reaction (RT-PCR) analysis of apoptotic genes revealed the upregulation of the p53 tumor suppressor gene, Bax, and caspase-9. The results assembled from this study further indicates that p44/42 MAPK, p53, caspase 9 and Bax play a major role in the mechanism of apoptosis in the MCF-7 breast cancer cells.
Keywords
AuNPs; Gold nanoparticles; MCF-7; Apoptosis; Gene expressions; Cancer
Abbreviations
MCF: Michigan Cancer Foundation; AuNPs: Gold Nanoparticles; ATCC: American Type Culture Collection; FBS: Fetal Bovine Serum; MTS: 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethyl phenyl)-2-(4-sulfophenyl)- 2H-tetrazolium); PBS: Phosphate Buffered Saline; M-PER: Mammalian Protein Extraction Reagent; BCA: Bicinchoninic acid; TBST: Tris Buffered Saline and Tween 20; MAPK: Mitogen-Activated Protein Kinase; IgG HRP: Immunoglobulin G Hormone Releasing Peptide; GAPDH: Glyceraldehyde 3-Phosphate Dehydrogenase
Introduction
Nanotechnology has become a very rampant research topic over the past years ranging from in vitro to in vivo clinical research using various metal and nonmetal particles targeting cancer cells and tumors [1]. One of the established advantages of the nanoparticles is its size and its potential to be programmed to specifically target organelles or tumors [2]. Gold is popularly known as an element on the periodic table with an oxidation number of +3 and +1. It has an atomic number of 79 and an atomic mass of 197 amu. It is a transition metal and is solid at room temperature. Gold is mostly naturally occurring but can also be synthesized, which is rare. Though gold has been used for a while in orthodontic medicine, electronics, and aeronautic engineering, a small size (nanoparticle) has been accepted by multiple researchers as a probable agent for cancer remedy in biomedicine [3]. A popular method for gold nanoparticle synthesis is a reduction reaction of tetrachloroauric acid (HAuCl4) by citrate commonly called the Turkeivich method. Other reducing agents such as plants as illustrated by Raghunandan et al. and Geetha et al. and fungi [4-6] have been used as well. Researchers have also reported how the possible sizes and shapes of synthesized gold nanoparticles [7]
Material and Methods
Cell culture
Cells were purchased from ATCC (American Type Culture Collection) and maintained in 5% DMSO at -80°C until use. For culture, cells were maintained in RPMI 1460 Medium (Gibco, CA) enriched with 10% FBS (Fetal Bovine Serum) and 1% antibiotic/ antimycotic at 37°C with 5% CO2 in a humidified atmosphere. Cells were plated at 5 × 104 cells/well to obtain a pattern of growth for further studies.
Synthesis of gold nanoparticles
The nanoparticles were synthesized using a modification of the Turkevich method by Cesbron et al. [11]. Briefly, 0.01 g of HAuCl4 was added to 50 ml of heated water. 1% of sodium citrate (Fisher Scientific) was then added to the heated stirring solution. A reduction reaction was noticed when the solution turned into a wine color. The solution was purified using a 0.2 μm filter, protected from light and stored at 4°C.
Treatment of cells
MCF-7 cells were seeded at 3 × 105 cells per plate, allowed to attach for 24 hours and incubated in a humidified atmosphere at 37°C with 5% CO2. The cells were treated with 0, 5, 10, 15 and 20 μg/ml of AuNPs in culture media. Cells were counted using the improved Neubauer hemocytometer chamber for 5 consecutive days.
Cell viability by MTS assay
The MCF-7 cells were plated at a density of 1 × 104 cells per well in a 96 well plate and allowed to attach. Thereafter, the cells were treated with AuNPs at 0, 5, 10, 15 and 20 μg/ml for 72 hours. Manufacturer’s (Promega) instructions were followed. Briefly, 20 μl of MTS solution was added to each well and incubated for 4 hours. The plates were read at 492 nm using a SkanIt plate reader. The data obtained were statistically analyzed.
Western blot analysis
The MCF-7 cells (5 × 105) were incubated in petri dishes and allowed to attach for 24 hours. The cells were treated with similar concentrations as stated above of AuNPs for three consecutive days, trypsinized using EDTA-Trypsin (Gibco), washed with PBS and stored at -20°C. Protein was extracted from the cells using M-PER (Mammalian Protein Extraction Reagent) (with protease Inhibitor) and was quantified using the BCA assay kit (Thermo Scientific). The samples (20 μg) of protein to be run were mixed with lysis buffer and loading buffer (dye) and put into the 10% SDS-PAGE (Nupage 10% Bis-Tris Gel, Invitrogen). The gel was run at 160V and then transferred to the nitrocellulose membrane (Bio-rad, 0.45μm) at 120V. The membrane was blocked using 5% blotting grade dry milk solution in 1x TBST for an hour and washed in the same buffer. Thereafter, the membrane was soaked overnight in p44/42 MAPK (1:1000) primary antibody. The membrane was washed three times with 1x TBST and soaked in a secondary antibody (Goat anti-Mouse IgG HRP 1:2000) for an hour. The membrane was then stripped and immersed in GAPDH primary antibody with a corresponding secondary antibody. After repetitive washing, the blot was soaked in chemiluminescence (Clarity Western ECL Substrate, Biorad) solution. Images were visualized using the ChemiDoc Imaging System and Software (Biorad).
Real-time quantitative polymerase chain reaction
The MCF-7 cells (5 × 105) were plated in petri dishes and allowed to attach for 24 hours. Cells were treated for 72 hours with similar concentrations as stated above of gold nanoparticles. RNA was extracted from the cells using the Aurum Total RNA Mini Kit (Biorad). Briefly, the cells were lysed using lysis solution supplemented with β-methcarpenol, repetitive washing with 60% ethanol, high and low stringency solution and DNase solution was used to extract the RNA in a binding column. Finally, the elution solution at 70ºC was used to collect the RNA extract for each sample. For the reverse transcription process, the RNA template was incubated for an hour at 42°C with iScript Reverse Transcription Supermix (Biorad) for cDNA synthesis. Using a SsoAdvanced Universal SYBR Green Supermix (Biorad), and 5 primers for each sample (p53, Caspase 9, BaCl2, Bax, and β-actin). PCR plate (Hard-Shell PCR Plates 96-well, thin-wall, Biorad) was set up and run for 40 cycles at 98°C (30 secs), 95°C (15 secs) and 60°C (25 secs) to obtain Ct values using a CFX96 instrument and software (Biorad).
Statistical Analysis
Statistical data for the reverse transcription-polymerase chain reaction was obtained in reference to β-actin. The method of calculation was obtained from Kannan [12]. The difference between the Ct values of subtracted and treated samples were obtained for all of the targeted genes (p53, caspase-9, bcl2, bax, and β-actin). The difference between these two newly obtained values was calculated to the negative power of two, for acquiring the relative fold change.
Results
Characterization and purification of gold nanoparticles
The synthesized AuNPs were determined using a UV-Vis Spectrophotometer measured at 525 nm wavelength. Though there are studies that have characterized gold nanoparticles by ligand adsorption [12], the AuNPs in this research were classified according to the Cytodiagnostics Charts [13] and a mathematical study conducted by Rahman [14]. The nanoparticle size was estimated to be about 35 nm size and an approximate concentration of about 50 μg/ml. The solution was purified using a 0.2 μm filter. Figure 1 showed the spectrum of the synthesized AuNPs from the UV-Vis spectrophotometer.
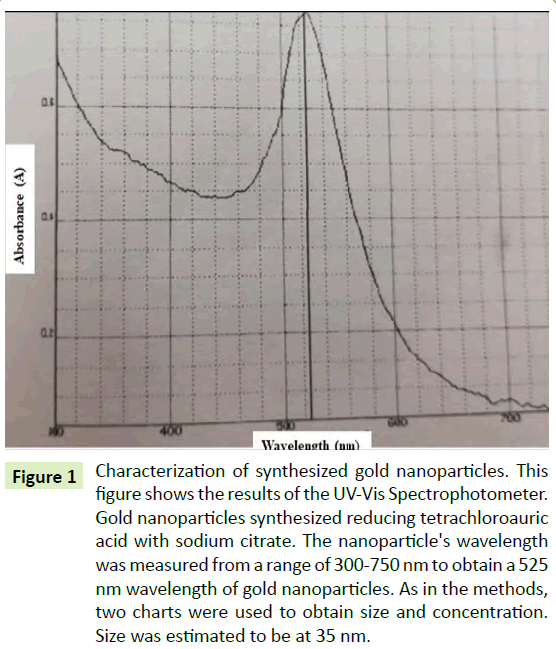
Figure 1: Characterization of synthesized gold nanoparticles. This figure shows the results of the UV-Vis Spectrophotometer. Gold nanoparticles synthesized reducing tetrachloroauric acid with sodium citrate. The nanoparticle's wavelength was measured from a range of 300-750 nm to obtain a 525 nm wavelength of gold nanoparticles. As in the methods, two charts were used to obtain size and concentration. Size was estimated to be at 35 nm.
Effects of gold nanoparticles on the viability of MCF-7 cells
Cells were treated with 0, 5, 10, 15 and 20 μg/ml of AuNPs. The data were collected daily for 5 days to understand the effects of AuNPs treatment on this cell line. Figure 2 showed viability of the cells at different concentrations of AuNPs for 5 consecutive days. As shown in Figure 2, the cell proliferation was decreased as concentration of AuNPs was increased. At 20 μg/ml of AuNP concentration, the cell viability was reduced by over 50% after 24 hours of treatment.
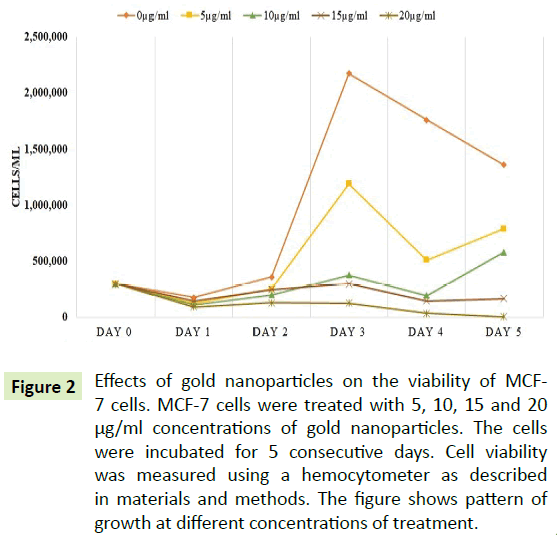
Figure 2: Effects of gold nanoparticles on the viability of MCF- 7 cells. MCF-7 cells were treated with 5, 10, 15 and 20 µg/ml concentrations of gold nanoparticles. The cells were incubated for 5 consecutive days. Cell viability was measured using a hemocytometer as described in materials and methods. The figure shows pattern of growth at different concentrations of treatment.
Inhibition of proliferation shown by MTS assay
To measure the cytotoxic effects of AuNPs, the cells were treated at various concentrations of AuNPs and the data was retrieved after 72 hours of treatment. Figure 3 showed that due to the increase in AuNPs concentration, there was a proportional increase in inhibition of cells according to absorbance. Compared with the control cells, the Figure 3 also showed 35, 50 and 90% inhibition of the cells at the concentrations of 5, 10 and 15 μg/ml of AuNPs, respectively.
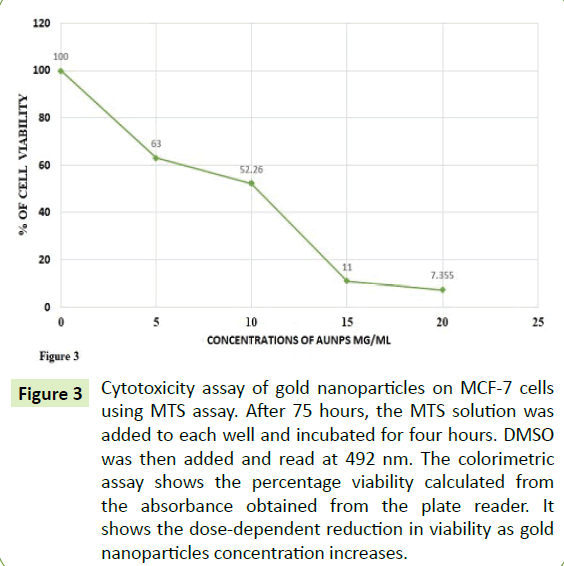
Figure 3:Cytotoxicity assay of gold nanoparticles on MCF-7 cells using MTS assay. After 75 hours, the MTS solution was added to each well and incubated for four hours. DMSO was then added and read at 492 nm. The colorimetric assay shows the percentage viability calculated from the absorbance obtained from the plate reader. It shows the dose-dependent reduction in viability as gold nanoparticles concentration increases.
Downregulation of p44/42 MAPK by Western blot analysis
To depict the effects of the AuNPs on the phosphorylated proteins p44/42 in the MAPK pathway, western blotting was performed. Figure 4 showed that there was a downregulation of p44/42 after treatment with AuNPs indicating the inhibition of the phosphorylation which is correlated with the impediment of cancer cell growth. This figure further showed a decrease in bandwidth by 50% compared to the control, as well as the consistency of the bandwidth of GAPDH as a house-keeping protein.
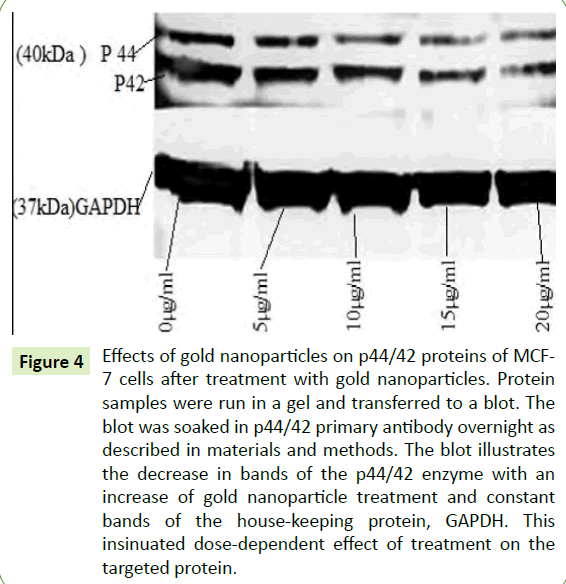
Figure 4:Effects of gold nanoparticles on p44/42 proteins of MCF- 7 cells after treatment with gold nanoparticles. Protein samples were run in a gel and transferred to a blot. The blot was soaked in p44/42 primary antibody overnight as described in materials and methods. The blot illustrates the decrease in bands of the p44/42 enzyme with an increase of gold nanoparticle treatment and constant bands of the house-keeping protein, GAPDH. This insinuated dose-dependent effect of treatment on the targeted protein.
Activation of p53, caspase-9, and Bax indicated by RT-qPCR
RT-PCR gives quantitative information about the expression of genes. Figure 5 showed the quantitative data of the gene expression by RT-PCR. Statistical computations were carried out using β-actin to obtain double delta Ct for each of the averages of the targeted gene samples. The Figure 5 showed a 9-fold upregulation of the p53 gene, which is a tumor suppressor gene, suggesting that there was a significant increase in p53 expression after treatment with AuNPs. The expression of caspase-9 gene was increased 3-fold, this indicating that there was significant caspase-mediated cell death due to the treatment with AuNPs. However, bax and bcl-2, both apoptotic regulators, were expressed to ~2.5-fold and ~0.6-fold, respectively. The Bax appears to be significantly upregulated, bcl-2 expressions were perceived to be insignificant.
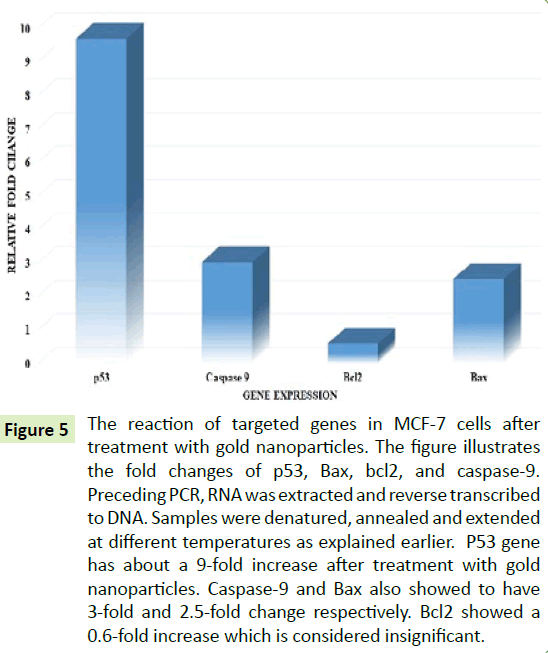
Figure 5:The reaction of targeted genes in MCF-7 cells after treatment with gold nanoparticles. The figure illustrates the fold changes of p53, Bax, bcl2, and caspase-9. Preceding PCR, RNA was extracted and reverse transcribed to DNA. Samples were denatured, annealed and extended at different temperatures as explained earlier. P53 gene has about a 9-fold increase after treatment with gold nanoparticles. Caspase-9 and Bax also showed to have 3-fold and 2.5-fold change respectively. Bcl2 showed a 0.6-fold increase which is considered insignificant.
Discussion
Nanoparticles are not novelle. Over the past decades there has been a high rise in the use of different kinds of nanoparticles. In 2012, cobalt oxide nanoparticles [15] were reported to induce apoptosis in Jurkat and KB cell lines in vitro studies. Aptamer labeled paclitaxel loaded poly lactic-co-glycolic acid nanoparticles were shown to decrease viability in GI-1 cancer cells [16]. Also, Rajeeth et al. [17] disclosed that synthesized silica nanoparticles increased the expression of p53 significantly in MCF-7 cells after analysis through western blotting. Though, only gold nanoparticles are used in this study, there seems to be a strong correlation in the activities of nanoparticles as remedies for cancer.
Most of the recent studies carried out have combined the gold nanoparticles with either light or other biological substances. In a study carried out by Bhowmik et al. [18] where the effects of gold nanoparticles with a toxin obtained from an Indian cobra on human lymphocytes and leukemic cells showed that there was induced apoptosis in leukemic cells. Vedantam et al. [19] reported that synthesized gold nanoparticles had a significant effect on prostate cancer cells (DU-145) after 2-3 days. They also indicated the effects of combined sugar D-mannan (Mn) with gold nanoparticles explaining that, there was twice as much uptake than merely gold nanoparticles on prostate cancer cells. Peptides have also been conjugated with gold nanoparticles by Kalmodia et al. [20] to be tested on Y79 retinoblastoma cells (RCB1645). They stipulated that both peptides indicated dose-dependent inhibition of reactive oxygen species and also showing there was more uptake when the gold nanoparticles were conjugated with the peptides. In a study where grapes were used to synthesize gold nanoparticles suggested that the plasma of the cancer cell (HBL- 100) was affected negatively and apoptosis was investigated [21]. Although the gold nanoparticles in this study were not combined nor enhanced, they demonstrated significant effects on the MCF- 7 cells. Therefore, it is which is crucial to highlight some of the approaches that have been taken using gold nanoparticles.
Several studies reported the activities of gold nanoparticles on a molecular aspect over the past years. These were concentrated on how gold nanoparticles could be responsible for reducing proliferation due to targeting organelles such as mitochondria, endoplasmic reticulum, nucleus, and the Golgi apparatus, lysosomes, proteasomes, etc. [22]. Epidermal growth factor has also been conjugated with gold nanoparticles to observe effects in vitro on the A549 lung cancer cell line. It was reported that the EGF conjugated AuNPs entered the cell’s cytoplasm [23]. Another study reported that a smaller size (1.9 nm) of gold nanoparticles suspended in sterile water has been reported to induce apoptosis in DU-145 cell line after expressions of apoptotic agents such as caspase-9 and cleaved PARP were analyzed by western blotting [24-26]. This study only focused on the expressions of a few signaling proteins in the MCF-7 cell line treated with citrate colloidal gold nanoparticle which created the need for our study to account for the expression of other proteins. A closely related research reported the expressions of most genes targeted in this study. In 2012, Selim et al. [10] reported that after treatment of MCF-7 cells with gold nanoparticles, the tumor suppressor gene p53, caspase-9, caspase-3, and Bax were significantly upregulated while bcl2 was downregulated. Our study also illustrates how p53, caspase-9, Bax, BCL-2 and p44/42 manifest in MCF-7 human breast adenocarcinoma cells after treatment with gold nanoparticles. Therefore, it can be inferred that the increase of p53, Bax, and caspase-9 and downregulation of p44/42 conducts a considerable part on the mechanism of inhibition by gold nanoparticles.
Conclusion
In conclusion, the present study confirmed that the viability of MCF-7 cells was decreased with an increase in the concentration of AuNPs. The investigations were carried out to study the effects of the cells’ signal transduction pathway after treatment with colloidal gold nanoparticles. These results revealed induced apoptosis and inhibition of MCF-7 cells proliferation through the gene expressions of p53, caspase-9, Bax and p44/42. This study encourages further research in vivo and prospective clinical trials using AuNPs as an anticancer drug.
References
- England CG, Huang JS, James KT, Zhang G, Gobin AM, et al. (2015) Detection of phosphatidylcholine-coated gold nanoparticles in orthotopic pancreatic adenocarcinoma using hyperspectral imaging. PLoS One 10: e0129172.
- Lee J, Chatterjee DK, Lee MH, Krishnan S (2014) Gold nanoparticles in breast cancer treatment: promise and potential pitfalls. Cancer Lett 347: 46-53.
- Dreaden EC, Alkilany AM, Huang X, Murphy CJ, El-Sayed MA (2012) The golden age: Gold nanoparticles for biomedicine. Chem Soc Rev 41: 2740-2779.
- Raghunandan D, Ravishankar B, Sharanbasava G, Mahesh DB, Harsoor V, et al. (2011) Anti-cancer studies of noble metal nanoparticles synthesized using different plant extracts. Cancer Nanotechnol 2: 57-65.
- Geetha R, Ashokkumar T, Tamilselvan S, Govindaraju K, Sadiq M, et al. (2013) Green synthesis of gold nanoparticles and their anticancer activity. Cancer Nanotechnol 4: 91-98.
- Chauhan A, Zubair S, Tufail S, Sherwani A, Sajid M, et al. (2011) Fungus-mediated biological synthesis of gold nanoparticles: Potential in detection of liver cancer. Int J Nanomedicine 6: 2305.
- Guo J, Rahme K, He Y, Li LL, Holmes JD, et al. (2017) Gold nanoparticles enlighten the future of cancer theranostics. Int J Nanomedicine 12: 6131-6152.
- Khanyile S, Masamba P, Oyinloye BE, Mbatha LS, Kappo AP (2019) Current biochemical applications and future prospects of chlorotoxin in cancer diagnostics and therapeutics. Adv Pharm Bull 9: 510-520.
- Jain S, Hirst DG, Sullivan JMO (2011) Gold nanoparticles as novel agents for cancer therapy. Br J Radiol 85: 101-113.
- Selim ME, Hendi AA (2012) Gold nanoparticles induce apoptosis in mcf-7 human breast cancer cells.Asian Pac J Cancer Prev 13: 1617-1620.
- Cesbron Y, Shaheen U, Free P, Lévy R (2015) TAT and HA2 facilitate cellular uptake of gold nanoparticles but do not lead to cytosolic localisation.Plos One 10: e0121683.
- https://bitesizebio.com/24894/4-easy-steps-to-analyze-your-qpcr-data-using-double-delta-ct-analysis/
- https://www.cytodiagnostics.com/store/pc/Introduction-to-Gold-Nanoparticle-Characterization-d3.htm
- Rahman S (2016) Size and concentration analysis of gold nanoparticles with ultraviolet-visible spectroscopy.Undergraduate Journal of Mathematical Modeling: One+Two: 7.
- Chattopadhyay S, Chakraborty SP, Laha D, Baral R, Pramanik P, et al. (2012) Surface-modified cobalt oxide nanoparticles: New opportunities for anti-cancer drug development.Cancer Nanotechnol 3:13-23.
- Aravind A, Varghese SH, Veeranarayanan S, Mathew A, Nagaoka Y, et al. (2012) Aptamer-labeled PLGA nanoparticles for targeting cancer cells.Cancer Nanotechnol 3: 1-12.
- Rejeeth C, Kannan S, Muthuchelian K (2012) Development of in vitro gene delivery system using ORMOSIL nanoparticle: Analysis of p53 gene expression in cultured breast cancer cell (MCF-7).Cancer Nanotechnol 3: 55-63.
- Bhowmik T, Saha PP, Dasgupta A, Gomes A (2013) Antileukemic potential of PEGylated gold nanoparticle conjugated with protein toxin (NKCT1) isolated from Indian cobra (Naja kaouthia) venom.Cancer Nanotechnol 4: 39-55.
- Vedantam P, Huang G, Tzeng TR (2013) Size-dependent cellular toxicity and uptake of commercial colloidal gold nanoparticles in DU-145 cells.Cancer Nanotechnol 4: 13-20.
- Kalmodia S, Vandhana S, Rama BR, Jayashree B, Seethalakshmi TS, et al. (2016) Bio-conjugation of antioxidant peptide on surface-modified gold nanoparticles: A novel approach to enhance the radical scavenging property in cancer cell.Cancer Nanotechnol 7.
- Amarnath K, Mathew NL, Nellore J, Siddarth CR, Kumar J (2011) Facile synthesis of biocompatible gold nanoparticles from Vites vinefera and its cellular internalization against HBL-100 cells.Cancer Nanotechnol 2: 121-132.
- Kodiha M, Wang YM, Hutter E, Maysinger D, Stochaj U (2015) Off to the organelles - killing cancer cells with targeted gold nanoparticles.Theranostics 5: 357-370.
- Silva CO, Petersen SB, Reis CP, Rijo P, Molpeceres J, et al. (2016) EGF functionalized polymer-coated gold nanoparticles promote EGF photostability and EGFR internalization for photothermal therapy.Plos One 11: e0165419.
- Coulter J (2012) Cell type-dependent uptake, localization, and cytotoxicity of 1.9 nm gold nanoparticles.Int J Nanomedicine 2012: 2673.
- Wallace HM (2013) Cancer therapy: Targeting mitochondria and other sub-cellular organelles. Curr Pharm Des 20: 201-222.
- Wang F, Li C, Cheng J, Yuan Z (2016) Recent advances on inorganic nanoparticle-based cancer therapeutic agents.Int J Environ Res Public Health 13: 1182.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences
